|
Note: the numbers in
square brackets refer (and link) to the List of
Publications
Fast Protein Folding Dynamics
a-Helix Folding
Dynamics
The first phase of protein folding is generally believed
to be the formation of secondary structural elements on the time scale of nano- to microseconds, which then act as nucleation
sites for the collapse to the native structure. Such fast folding processes
can be observed using temperature jumps, induced by a nanosecond laser
pulse, which heats the solvent to a temperature above the transition tem≠pe≠ra≠ture
of the protein. The kinetics of the ensuing unfolding can then be observed
by time-resolved IR-spec≠tros≠copy.
Several
novel aspects of helix folding have been studied:
* Bulky
side chains have been shown not to slow down folding [22].
* The substitution of individual amino acid
residues could be shown to affect not only helical stability, but also the
dynamics of the helix-coil relaxation [24].
*
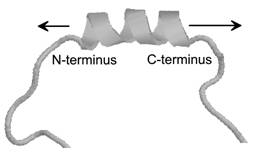
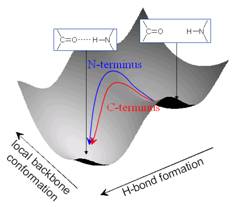
Isotope-edited time-resolved IR spectroscopy, which allows the observation
of helix dynamics at the residue level, was used to show that the helix
dynamics at the C-terminus of an a-helix are faster than those at the N-terminus [23], most likely because the backbone carbonyls of the last three
residues at the C-terminus are not hydrogen bonded and thus allow for
greater flexibility.
 †††††††††††††††††††† †††††††††††††††††††† 
* The
effect of solvent conditions, such as pH or salt content were
investigated in detail [25].
* The
dependence of helix folding dynamics on the overall helicity
of the peptide, which can be modified by pH (in polyglutamic
acid) [32].
In
addition to IR spectroscopy, we also were involved in the development of
ns-time resolved CD spectroscopy as an alternative detection method for
temperature jump-induced helix folding dynamics, which provides the
advantage of yielding absolute values of the helical content of a sample [28].
More
recent, and as yet unpublished results, focus on the folding dynamics of
the coiled-coil motif, which consists of two helices which wrap around each
other.
Fast
Protein Folding Triggers
The
absence of fast trigger methods for protein folding is one of the main
obstacles for investigating the dynamics of the first steps of this process
[21]. We have shown that photorelease of
a nitrobenzyl group from the backbone of a
modified peptide occurs on the nanosecond time scale, and thus could be
used as fast trigger mechanism for protein folding [Abstract17]. Work on other trigger mechanisms is in preparation.
Roughness
of the Protein Energy Landscape
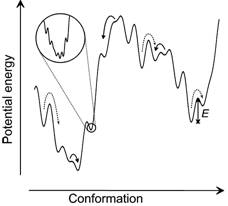
The recombination dynamics of a UV-photolyzed
non-native disulphide bond in a protein proves that the polypeptide
backbone undergoes anomalous backbone diffusion behaviour [34,
31,
13]. Analysis of
these data shows that the roughness of the proteinís energy landscape is of
the order of 4-5 kBT.
Coil/Globule
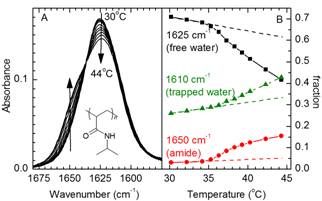
Transition of pNIPAm
poly(N-isopropylacrylamide)
collapses from an extended coil state to a globular conformation when
heated above the Lower Critical Solution Temperature or upon changes in pH,
with a wide range of potential practical applications. We have investigated
this coil/globule transition by FTIR spectroscopy and temperature jump/IR
methods. The results indicate trapped water molecules inside pNIPAm globules and show structural changes occurring
on the time scale of seconds in single chains of pNIPAm,
whereas cross-linked pNIPAm shows much faster
structural changes [Book12].
Stabilization
of Insulin on Hydrophobic Surfaces
Insulin is prone to amyloid fibril
formation, especially under the technologically important acidic conditions
and in the presence of hydrophobic surfaces. Using ATR-FTIR and
sum-frequency spectroscopy, we were able to show that even under these
conditions insulin adsorbs in its native structure to both hydrophilic and
hydrophobic surfaces, but converts to amyloid-like
structures at elevated temperatures on hydrophobic surfaces [35].
Structure
of Protein Variants
FTIR spectroscopy has been used to investigate the internal
structure of different variants of the titin Z1
domain to show that its core structure remains intact upon the introduction
of recognition sequences in a surface loop [30].
|