Cell Signalling Laboratory
| |
| Home |
| Selected publications |
| Group Members |
| Links |
| Vacancies |
| COMPARTMENTALISATION |
The cell consists of discrete compartments (organelles) such as the nucleus, Golgi, endoplasmic reticulum, plasma membrane, endosomes and mitochondria that are separated from each other by a protein rich soup called cytosol. Each organelle specialises in a different function by concentrating different groups of proteins and lipids together and regulating their interactions.
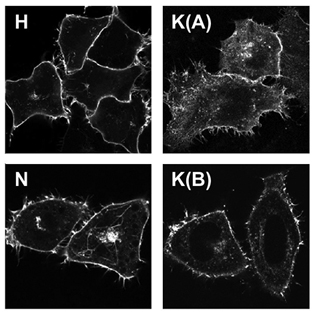
1. The cell surface consists of a mosaic of microdomains that cluster different pools of proteins and lipids into tiny (<20 nanometer) patches visible only using electron microscopy or advanced light microscopy techniques. The different Ras isoforms do not appear to co-localise in these patches; a model that we are testing is that these different Ras microdomains will have different pools of signalling proteins preferentially co-localised allowing each type of microdomain to generate a different output.
2. Ras isoforms are found on internal organelles to different extents. For example, N-Ras is often more strongly associated with the Golgi complex (bright central spot in images below) than the other isoforms. This means that it is likely to be better at activating pathways localised on this organelle than the other isoforms.
Latest
news Group members Key collaborators Productive long term interactions exist with the Royle lab investigating mitotic spindle assembly and with the Brust lab developing novel biofunctionalised nano materials.
. |