
Requirements: Candidates should be enthusiastic and ambitious and have some basic knowledge of statistics and its application to biological data. Knowledge in programming languages such as R, python, perl, C or other would be preferred but are not required. Training in computational biology, in particular predictive biology, will be provided. As this project encapsulates a strong experimental component it is advised that the student should have laboratory experience, ideally already have experience running TG236, and have worked with environmental species of interest.
DEVELOPING ADVERSE OUTCOME PATHWAYS FROM EXPOSURE DYNAMICS IN ZEBRAFISH EMBRYOS
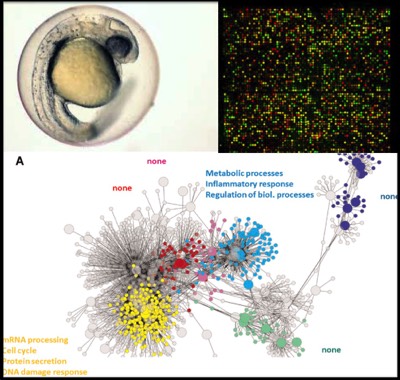
Background: Adverse outcome pathways (AOP's) are the conceptualization of knowledge into logical chains of events linking molecular initiating events through key processes to adverse outcomes. Their applicability to regulatory and monitoring processes has been demonstrated, however the development and discovery of AOP's still proves to be a challenge. To address this, data-driven approaches can leverage the power of computational biology and high dimensional data sets to accelerate the discovery of AOP's. Using the newly established OECD test-guideline 236, the fish embryo toxicity test (FET), the successful candidate will develop a large molecular dose response dataset and expand the current EPIC-map and develop quantitative AOP's.
The successful candidate will have the opportunity of working closely with CEFAS, learning from their expertise in zebrafish biology and its application to regulation and environmental monitoring. Experiments with zebrafish embryos will be conducted at the CEFAS Weymouth laboratory. The successful candidate will also have the opportunity of working with AstraZeneca and utilising their expertise in AOP's and their application in industry.
Training: This project will provide diverse training in computational biology approaches and associated programming languages (especially R and its libraries and functions). In addition the student will learn how to use data-driven approaches and generate and validate hypotheses high dimensional data resources. The student will also gain more generic training from the IIB PhD training programme and support through both infrastructure and access to expertise in molecular biology, ecotoxicology and computational biology.
Send an e-mail to Philipp Antczak or Stew Plaistow if you have any questions or are interested in applying.
Applications are accepted all year round. Applications (CV, letter of application, 2 referees) by email to biolres@liv.ac.uk. Interviews will be arranged as required.