Angewandte Chemie International Edition, 49 (2010) 2344–2348
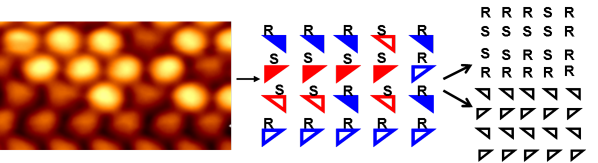
Amino acids find their feet: Scanning tunneling microscopy of racemic (R,S)-proline on Cu(110) reveals rows of random chiral amino acid sequences, showing that organization is not governed by molecular chirality. Instead, the system is dictated by a strict heterochiral adsorption-footprint template, in which each adsorption position can be occupied by either enantiomer (see picture), resulting in a random solid solution in 2D.

We present the first molecule-by-molecule mapping of a racemic amino acid system at a surface, showing the structure of the (4 × 2) monolayer of (R,S)-proline on Cu(110) is driven, not by molecular chirality, but by the chirality of the adsorption footprint. This situation leads to a random arrangement of molecular chirality but results in a strict heterochiral adsorption-footprint pattern. This effectively creates a 2D random solid solution, which is a rare occurrence in 3D crystals but, interestingly, may become more prevalent in 2D owing to surface interactions, as shown herein. For amino acids, this behavior is a departure from that found in 3D where they crystallize as racemic compounds. Our work provides an important advance into understanding amino acid ordering at surfaces, with insights provided by STM, low energy electron diffraction (LEED), reflection absorption infrared spectroscopy (RAIRS), and periodic density-functional theory (DFT).
Copyright © 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. Copies of the full text are available on request